| Transcript | Length (nt) | A | C | G | T |
|---|---|---|---|---|---|
| NM_001317214.3 | 3407 | 28.53% | 22.07% | 22.04% | 27.36% |
| NM_033646.4 | 12126 | 33.24% | 17.33% | 18.51% | 30.92% |
| NM_004361.5 | 12136 | 33.03% | 17.52% | 18.68% | 30.77% |
| NM_001362438.2 | 12938 | 32.13% | 18.00% | 19.49% | 30.38% |
1 Sequence Retrieval and Analysis of Human Cadherin-7
2 Introduction
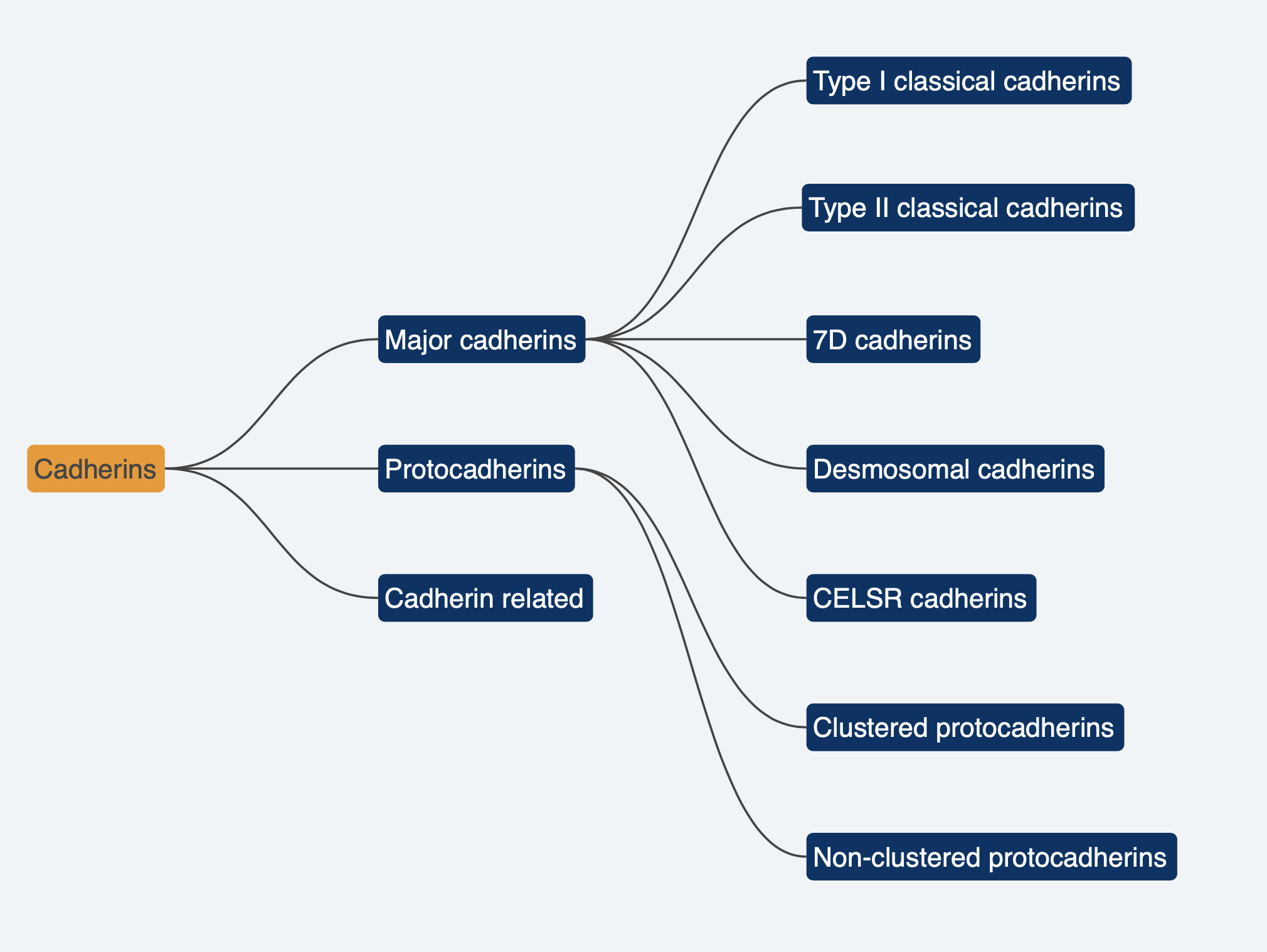
Cadherins represent a superfamily of proteins vital for mediating cell-cell adhesion and intracellular signalling [1]. These proteins are integral to maintaining tissue integrity, regulating cellular behavior, and governing fundamental cellular functions. Our research focuses on human Cadherin-7 (CDH7). CDH7 is encoded by a gene located within a gene cluster on chromosome 18 and belongs to the type II classical cadherin category (Figure 1)[2].
The research aim is the retrieval and analysis of mRNA and protein sequences of human CDH7 to gain insights into structural and functional differences. We aim to explore the implications of alternative splicing of human CDH7 and how they fit into the broader context of the Cadherin protein family.
3 Data & Methods
Various data sources were used to retrieve essential information. We obtained mRNA sequences from the NCBI Nucleotide database [4] and protein sequences from the NCBI Protein database [5]. The HGNC gene name database was utilised to obtain standardised gene symbols, most crucially the Cadherin group data which contains all gene symbols for the entire Cadherin family of proteins [3].
Our research methodology can be divided into several key steps:
- Data Retrieval: We used a combination of Biopython [6] and its Entrez module to retrieve and analyse mRNA and protein sequences from NCBI. In cases where programmatic data retrieval did not offer clear advantages, we resorted to using web interfaces, such as for transcript tables. We applied appropriate NCBI filters and query parameters, following the guidelines available at NCBI. Where possible, staff-curated and RefSeq sequence data was used. Relevant queries and databases are documented in Table 11.
- Alignment Algorithms: To perform sequence alignments, we utilised a combination of tools, including Biopython’s pairwise module with global (Needleman-Wunsch [7]) and local (Smith-Waterman [8]) alignment algorithms. Additionally, we employed BLAST [9] for protein and nucleotide sequence comparisons using the blastp and blastn algorithms with the compositional score matrix adjustment method [10].
- Reproducibility of this Report: Ensuring the reproducibility of our research was a top priority. We achieved this through several means: 1) Versioned Accession Numbers: We provided versioned accession numbers to ensure that data can be traced accurately. 2) Quarto Document Production: This report was generated using Quarto [11], a tool that mandates code compilation for document compilation. This ensures that the code remains synchronised with the document and is always executable.
Detailed methods are provided when results are presented to enhance the coherence of this report.
4 Errata
Part 2:
- Global alignment would have been better, this one is deceptively neat.
- The number of determined exons for human CDH-7 is 12, not 15.
- Part of the transmembrane domain, this is crucial because it is no longer long enough to span the membrane.
- Missing part analysis: while indeed the missing part makes it not be a transmembrane protein (i.e. it makes it soluble) it still is functional in the sense it binds to other, full cadherins, regulating their their function through inhibition.
Part 3:
- This is only a part of the ~245 that human proteins with a Cadherin Domain.
- Second part: should be CDH-20, CDH-18 and CDH-10.
5 Results
5.1 Part One – Retreiving and Analysing mRNA and Protein Sequences of Human Cadherin-7
Cadherin-7 (CDH7) is a calcium-dependent cell-cell adhesion molecule. It undergoes alternative splicing, resulting in the production of various mRNA transcript variants and protein isoforms [12]. The general protein structure of CDH7 is characterised by five extracellular cadherin repeats, a transmembrane region, and a cytoplasmic tail, which collectively facilitate its adhesive and signalling functions [12].
Research has linked CDH7 to a range of diseases, positioning it as a potential risk factor in various pathological conditions. This includes bipolar disorder [13], melanoma cancer [2], and major depressive disorder [14]. These findings emphasise the significance of CDH7 in both neurological and oncological research.
In the initial phase of our research, we employed query \(Q1\) from Table 11 to identify and retrieve relevant transcripts. We used the resulting accession numbers to obtain the respective GenBank file and to retrieve the transcript variant sequences, from which length (in nucleotides, nt) and nucleotide composition were computed (Table 1).
The next step was to extract the coding sequence (CDS) from the records and use Biopython’s translation capability to obtain the protein isoform sequences, resulting in Table 2.
| Source transcript | Protein length | Highest count amino acid (count) |
|---|---|---|
| NM_001317214.3 | 630 | S (52x) |
| NM_001362438.2 | 785 | S (66x) |
| NM_004361.5 | 785 | S (66x) |
| NM_033646.4 | 785 | S (66x) |
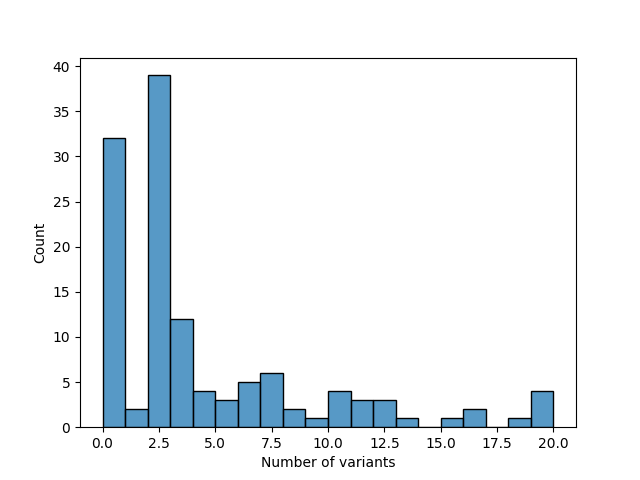
As additional piece of analysis, we investigated which other cadherin genes undergo alternate splicing by estimating the number of transcript variants present in the NCBI nucleotide database. First, we obtained all official gene symbols of the Cadherin family from HGNC, as stated in Section 3. For each of the symbols, we obtained the GenBank file and checked if it contained the string “transcript variant”. We further refined the investigation to prefer curated RefSeq database entries to predicted ones and detect if two search results have a different accession numbers but refer to the same sequence (may happen when one of the entries is outdated / was predicted but then confirmed by NCBI staff). Filtering by accession number syntax [15] was utilised to achieve this. The results are documented in Table 3 and Table 14.
| HNGC ID | Number of variants |
|---|---|
| PCDH15 | 20 |
| RET | 20 |
| PCDH11X | 20 |
| CDH18 | 20 |
| CDHR5 | 18 |
Figure 2 shows the distribution of the number of transcript variants for each Cadherin gene. The majority of Cadherin genes have 2-4 or no transcript variants.
5.2 Part Two – Comparing mRNA transcripts & protein isoforms of Human Cadherin-7 and interpretting the consequences of alternate transcription
We conducted pairwise sequence comparisons between the longest and shortest human CDH7 mRNA transcripts (NM_001362438.2 and NM_001317214.3, respectively). To accomplish this, we employed the Biopython pairwise2 module and compared the coding sequences of the transcripts. This choice was driven by the fact that the coding sequence encodes the protein. The parameters and details of the comparison are listed in Table 4.
| Alignment method | Match | Mismatch | Gap opening penalty | Gap extension penalty |
|---|---|---|---|---|
| Local | 1 | -1 | -3 | -1.5 |
The parameters selection was influenced by the expectation of high similarity between the sequences. Given that the transcripts belong to the same gene within the same species, we anticipated a high degree of sequence conservation. To achieve this, we chose parameters that penalise gaps and mismatches significantly, aiming for a more accurate and stringent alignment.
We obtained a single high-scoring alignment with a length of 2358 and a bit-score of 1864. Remarkably, the two sequences align with 100% identity for the entire length of the shorter sequence. The shorter sequence is missing the entire end region of the longer sequence, suggesting that the differences between these two CDH7 mRNA transcripts are concentrated in the terminal region of the longer sequence.
| Matrix | Alignment method | Match | Mismatch | Gap open. penalty | Gap ext. pen. |
|---|---|---|---|---|---|
| Blosum90 | Local | 1 | -1 | -3 | -1.5 |
The next step of our research was to perform a comparison between the longest and shortest human CDH7 protein isoforms. While the annotation in the gene card for human CDH7 states that the longest protein isoform is encoded by NP_004352.2 [16], we found that the longest proetin isoform is encoded by three of the four transcript variants and not only one, so this is most likely an annotator’s faux pas. We chose protein isoform sequence NP_004352.2 for our analysis, knowing it encodes the longest isoform, comparing it to the shortest isoform sequence (NP_001304143.1).
The parameters for this sequence comparison (Table 5) were selected based on our hypothesis, which was that the sequences should align perfectly at the start and then diverge towards the end. The shorter protein isoform was expected to lack a significant portion of the end region present in the longer protein isoform.
We obtained 21 high-scoring alignments. The top-scoring alignment had a length of 785, which is the same as the length of the longer protein isoform, and a bit-score of 5292.40. As anticipated from our earlier coding sequence comparison, the two sequences aligned with high identity for the entire length of the shorter sequence, only diverging towards the end. Notably, the shorter sequence lacked almost the entire end region found in the longer sequence.
5.2.1 How many exons does the human CDH7 gene have?
Determining the precise number of exons in the human CDH7 gene can be a complex task, as different sources may provide varying numbers of exons due to differences in gene file versions and data sources. To establish an accurate count of exons on our own account, we followed a systematic strategy:
- Obtaining Exon Regions for Each Transcript Variant: We acquired exon regions for each transcript variant of the CDH7 gene from the annotations in the NCBI Nucleotide database’s GenBank files.
- Counting Exons: To count exons, we established a criterion that considered two regions as the same exon if they contained identical sequences, even if their start and end locations were different in their respective source transcripts.
The result is reported in Table 6 which shows that using our strategy, the human CDH7 gene has 15 exons.
To validate our findings, we compared our count of exons with information from other sources: The NCBI GenBank RefSeq annotation indicates 13 exons [16]. Our hypothesis is that this might be because some of the exons (e.g. exon_06 and exon_13), while differing in length, are highly similar and convey the same functionality, thus counted as the same exon by NCBI staff. The EBI search tool by the European Bioinformatics Institute, querying “CDH7” in Ensembl [17], reports 18 exons for human CDH7. This is likely due to the fact that Ensembl might contain different versions of the gene file and transcripts, with different exon annotations.
These variations highlight the complexities involved in exon counting due to differences in annotation methods and versions.
| Exon identifier | Length (nt) | Contained in (acc) |
|---|---|---|
| exon_01 | 118 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] |
| exon_02 | 120 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] |
| exon_03 | 122 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] |
| exon_04 | 129 | [‘NM_033646.4’] |
| exon_05 | 137 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] |
| exon_06 | 139 | [‘NM_004361.5’, ‘NM_001317214.3’] |
| exon_07 | 168 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] |
| exon_08 | 188 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] |
| exon_09 | 252 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001362438.2’] |
| exon_10 | 254 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] |
| exon_11 | 295 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] |
| exon_12 | 406 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] |
| exon_13 | 941 | [‘NM_001362438.2’] |
| exon_14 | 1460 | [‘NM_001317214.3’] |
| exon_15 | 9937 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001362438.2’] |
Table 6 enabled us to quickly identify which exons are contained in which transcript vairants. Recalling the shortest and longest transcript variants from Table 1, NM_001317214.3 and NM_001362438.2, we identified that the shortest transcript is missing exon_09, exon_13, and exon_15.
Interestingly, we observed that exon_06, which is present in the shortest transcript but not in the longest, is fully contained in exon_13. This suggests the possibility that these exons may encode similar functionalities and could be counted as the same exon.
Exon_09 and exon_15, the other two missing exons in the shortest transcript, are positioned adjacent to each other in their source transcript and are located at the end of the coding sequence.
The next step in our research was to explore the functional consequences of the missing parts in the protein encoded by the shortest transcript variant of the CDH7 gene. We conducted a Blastp analysis of the shortest transcript variant against the SwissProt database, filtering for human (Homo sapiens) and using the standard parameter settings enabled on NCBI.
The first result is the most significant one with a 100% identity match and e-value \(0.0\) for protein sequence Q9ULB5.2, which is the CDH7 isoform 1 preproprotein, the long protein isoform encoded by CDH7 [18]. The alignment results clearly indicate that the cytoplasmic region of the protein is missing in the shorter isoform.
The cytoplasmic region includes the C-terminal catenin-binding domain, which is responsible for binding to proteins like p120, beta-catenin, and plakoglobin [19]. Of particular importance is the interaction with beta-catenin, which binds to the cytoplasmic portion of cadherins as part of adherens junctions. Generally, the removal of the transmembrane anchoring domain, which can produce a soluble protein isoform, may have further functional implications. Such alternative splicing events can lead to a gain or loss of function, and the loss of membrane anchorage may disrupt the normal functioning of the protein [20]. The absence of this cytoplasmic region in the shorter CDH7 protein isoform suggests potential changes in cell adhesiveness and adherence junction formation. Beta-catenin is known to play a crucial role in preventing the proteasomal destruction of cadherin, ensuring the delivery of cadherin-beta-catenin complexes to the cell surface [19]. The loss of this interaction may have consequences for the adhesive properties of cells and their interactions with neighboring cells.
5.3 Part Three – Do other similar human Cadherin genes use alternate splicing in a similar way to Cadherin-7?
To identify all human proteins that contain a Cadherin domain we utilised the Cadherin gene group data from HGNC (see Section 3). For each Cadherin symbol, we performed a search in the NCBI protein database using query \(Q2\) (Table 11). In cases where multiple protein isoforms existed for the same Cadherin gene, we chose the longest isoform as the representative protein. Our research yielded a list of \(115\) human proteins that contain a Cadherin domain (Table 7, Table 13, [115 count merges duplicates]).
| HNCB Symbol | All proteins (acc) | Selected protein (acc) | |
|---|---|---|---|
| 0 | CDH16 | [‘13431340’] | 13431340 |
| 1 | CDH17 | [‘308153624’] | 308153624 |
| 2 | CDHR1 | [‘166980558’] | 166980558 |
| 3 | CDHR2 | [‘209572658’] | 209572658 |
| 4 | CDHR3 | [‘74758833’] | 74758833 |
Based on the accession numbers reported in Table 13, we programmatically retrieved all isoform sequences using Biopython, and performed pairwise comparison with the human CDH7 isoform sequence to obtain the three closest proteins. The parameters used for the pairwise comparison are reported in Table 8; the use of a stricter scoring matrix (BLOSUM90) was motivated by the expectation of a high degree of similarity between the sequences as they belong to the same gene family. Table 9 lists the top three closest Cadherin proteins to human CDH7.
| Alignment type | Match | Mismatch | Gap opening penalty | Gap extension penalty | Scoring matrix |
|---|---|---|---|---|---|
| local | 1 | -1 | -1 | -0.1 | BLOSUM90 |
| matrix | Bit score | HGNC Symbol | Protein (acc) | |
|---|---|---|---|---|
| 107 | BLOSUM90 | 3147.2 | CDH10 | 116241276 |
| 12 | BLOSUM90 | 3140.2 | FAT4 | 172046149 |
| 110 | BLOSUM90 | 3101 | CDH18 | 3023435 |
To investigate if the three closest Cadherin genes to CDH7 had short protein isoforms similar to the short protein isoform of CDH7, we selected these genes using HGNC symbols and accessed their gene cards on the NCBI web interface. From the transcript tables linked from the gene cards, we obtained the accession numbers of the shortest protein isoforms: ‘NP_078858.4’ for FAT4, ‘NP_001304151.1’ for CDH10, and ‘NP_001336491.1’ for CDH18.
We then compared these shortest isoforms to the short protein isoform of CDH7 to assess their similarities. The parameters used in the comparison were consistent with those documented in the previous parameters table (Table 8), accommodating moderate to high similarity without harsh gap penalties.
| Bit score | Protein (acc) | |
|---|---|---|
| 0 | 2589.1 | NP_078858.4 |
| 1 | 599.9 | NP_001304151.1 |
| 2 | 1573.7 | NP_001336491.1 |
The results of the pairwise comparisons are reported in Table 10. Looking at the alignments, we observed that all three protein isoforms do not feature the cytoplasmic region of the longest isoform, similar to the short isoform of CDH7. However, the results were not conclusive, as the alignments were not as high-scoring as expected. This may be due to the fact that the sequences are not as similar as expected, or that the scoring matrix and gap penalties were too relaxed, and we could have identified a different set of proteins as the most similar one. At this point, we concluded our research, acknowledging that we had reached the limits of our current resources and were uncertain about the interpretation of the results.
- Reported mutation:
- rs2291343
- https://www.nature.com/articles/ng1407
- Get mutated sequence, and normal CDH7, put into blast
- Or just blast normal cdh7 seq
6 Discussion
This research yielded a number of interesting insights into the human CDH7 gene and its protein isoforms. We identified that the shortest transcript variant of CDH7 is missing the end region of the longest transcript variant, and that the shortest protein isoform of CDH7 is missing the cytoplasmic region of the longest protein isoform. We also identified the exons and their locations for each transcript variant of CDH7, along with which exons are missing in the shortest transcript variant. The findings led to our hypothesis that CDH7 might lack beta-catenin binding capabilities, which may have consequences for the adhesive properties of cells associated with CDH7, and their interactions with neighboring cells.
We further conducted an exhaustive database search to find \(115\) human proteins that have a Cadherin domain. Given our chosen sequence comparison parameters, we found the three closest proteins to CDH7, which are isoforms of the FAT4, CDH10 and CDH18 genes.
We compared the shortest of those to the shortest CDH7 protein isoform, and found that they are all missing the cytoplasmic region of the longest isoform, like the shortest CDH7 isoform. However, our results were not conclusive and we were not sure how to interpret them, so we did not pursue this further but provided enough material to reproduce our results and continue the research from there.
Overall, our findings suggest that alternate splicing in the Cadherin family leads to varied protein functions. This is an interesting finding when set in relation to genetic diseases associated with Cadherin gene mutations, such as the CDH7 gene.
References
Appendix
| ID | Usage | Query structure | Queried Database |
|---|---|---|---|
| Q1 | Find RefSeq mRNA transcripts of a given gene for the human organism. The [TI] is a workaround to make up for the missing [Preferred Symbol] | {hgnc_symbol}[GENE] AND human[ORGN] AND mRNA[FILTER] AND RefSeq[FILTER] AND {hgnc_symbol}[TI] | NCBI nucleotide database |
| field in the API. | |||
| Q2 | Find protein for given gene for the human organism, filtered for swissprot data subset. | {hgnc_symbol}[GENE] AND human[ORGN] AND siwssprot[FILTER] | NCBI protein database, swissprot subset |
| Q3 | Find the human gene card for a given gene. | {hgnc_symbol}[GENE] AND human[ORGN] | NCBI gene database |
| Exon identifier | Length (nt) | Contained in (acc) | Start pos, respectively | End pos |
|---|---|---|---|---|
| exon_01 | 118 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] | [1820, 1830, 1830, 2632] | [1937, 1947, 1947, 2749] |
| exon_02 | 120 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] | [831, 841, 841, 1643] | [950, 960, 960, 1762] |
| exon_03 | 122 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] | [1698, 1708, 1708, 2510] | [1819, 1829, 1829, 2631] |
| exon_04 | 129 | [‘NM_033646.4’] | [1] | [129] |
| exon_05 | 137 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] | [1561, 1571, 1571, 2373] | [1697, 1707, 1707, 2509] |
| exon_06 | 139 | [‘NM_004361.5’, ‘NM_001317214.3’] | [1, 1] | [139, 139] |
| exon_07 | 168 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] | [951, 961, 961, 1763] | [1118, 1128, 1128, 1930] |
| exon_08 | 188 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] | [1119, 1129, 1129, 1931] | [1306, 1316, 1316, 2118] |
| exon_09 | 252 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001362438.2’] | [1938, 1948, 2750] | [2189, 2199, 3001] |
| exon_10 | 254 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] | [1307, 1317, 1317, 2119] | [1560, 1570, 1570, 2372] |
| exon_11 | 295 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] | [536, 546, 546, 1348] | [830, 840, 840, 1642] |
| exon_12 | 406 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001317214.3’, ‘NM_001362438.2’] | [130, 140, 140, 942] | [535, 545, 545, 1347] |
| exon_13 | 941 | [‘NM_001362438.2’] | [1] | [941] |
| exon_14 | 1460 | [‘NM_001317214.3’] | [1948] | [3407] |
| exon_15 | 9937 | [‘NM_033646.4’, ‘NM_004361.5’, ‘NM_001362438.2’] | [2190, 2200, 3002] | [12126, 12136, 12938] |
| HNCB Symbol | All proteins (acc) | Selected protein (acc) | |
|---|---|---|---|
| 0 | CDH16 | [‘13431340’] | 13431340 |
| 1 | CDH17 | [‘308153624’] | 308153624 |
| 2 | CDHR1 | [‘166980558’] | 166980558 |
| 3 | CDHR2 | [‘209572658’] | 209572658 |
| 4 | CDHR3 | [‘74758833’] | 74758833 |
| 5 | CDHR4 | [‘206558199’] | 206558199 |
| 6 | CDHR5 | [‘296439399’] | 296439399 |
| 7 | DCHS1 | [‘20139065’] | 20139065 |
| 8 | DCHS2 | [‘1890389199’] | 1890389199 |
| 9 | FAT1 | [‘334302792’] | 334302792 |
| 10 | FAT2 | [‘296434503’] | 296434503 |
| 11 | FAT3 | [‘1476413354’] | 1476413354 |
| 12 | FAT4 | [‘172046149’] | 172046149 |
| 13 | CLSTN1 | [‘23396490’] | 23396490 |
| 14 | CLSTN2 | [‘296434469’] | 296434469 |
| 15 | CLSTN3 | [‘23396507’] | 23396507 |
| 16 | PCDH15 | [‘116242702’] | 116242702 |
| 17 | RET | [‘547807’] | 547807 |
| 18 | CDH23 | [‘408359994’] | 408359994 |
| 19 | CELSR1 | [‘22095551’] | 22095551 |
| 20 | CELSR2 | [‘22095550’] | 22095550 |
| 21 | CELSR3 | [‘229462826’] | 229462826 |
| 24 | PCDHAC1 | [‘143811434’] | 143811434 |
| 25 | PCDHAC2 | [‘13878435’] | 13878435 |
| 26 | PCDHA1 | [‘13878434’] | 13878434 |
| 27 | PCDHA2 | [‘13878430’] | 13878430 |
| 28 | PCDHA3 | [‘13878429’] | 13878429 |
| 29 | PCDHA4 | [‘13878424’] | 13878424 |
| 30 | PCDHA5 | [‘13878428’] | 13878428 |
| 31 | PCDHA6 | [‘13878423’] | 13878423 |
| 32 | PCDHA7 | [‘13878422’] | 13878422 |
| 33 | PCDHA8 | [‘13878427’] | 13878427 |
| 34 | PCDHA9 | [‘13878426’] | 13878426 |
| 35 | PCDHA10 | [‘13878433’] | 13878433 |
| 36 | PCDHA11 | [‘13878432’] | 13878432 |
| 37 | PCDHA12 | [‘13878425’] | 13878425 |
| 38 | PCDHA13 | [‘13878431’] | 13878431 |
| 41 | PCDHB1 | [‘205371811’] | 205371811 |
| 42 | PCDHB2 | [‘13431378’] | 13431378 |
| 43 | PCDHB3 | [‘1714598584’] | 1714598584 |
| 44 | PCDHB4 | [‘13431376’] | 13431376 |
| 45 | PCDHB5 | [‘2203400475’] | 2203400475 |
| 46 | PCDHB6 | [‘2528974254’] | 2528974254 |
| 47 | PCDHB7 | [‘13431373’] | 13431373 |
| 48 | PCDHB8 | [‘1835921839’] | 1835921839 |
| 49 | PCDHB9 | [‘2203400717’] | 2203400717 |
| 50 | PCDHB10 | [‘13431371’] | 13431371 |
| 51 | PCDHB11 | [‘13431383’] | 13431383 |
| 52 | PCDHB12 | [‘13431382’] | 13431382 |
| 53 | PCDHB13 | [‘13431381’] | 13431381 |
| 54 | PCDHB14 | [‘13431380’] | 13431380 |
| 55 | PCDHB15 | [‘13431379’] | 13431379 |
| 56 | PCDHB16 | [‘2152360329’] | 2152360329 |
| 58 | PCDHB18P | [‘803341881’] | 803341881 |
| 61 | PCDHGA1 | [‘37999844’] | 37999844 |
| 62 | PCDHGA2 | [‘37999841’] | 37999841 |
| 63 | PCDHGA3 | [‘296439266’] | 296439266 |
| 64 | PCDHGA4 | [‘1248281085’] | 1248281085 |
| 65 | PCDHGA5 | [‘37999838’] | 37999838 |
| 66 | PCDHGA6 | [‘37999837’] | 37999837 |
| 67 | PCDHGA7 | [‘37999836’] | 37999836 |
| 68 | PCDHGA8 | [‘37999835’] | 37999835 |
| 69 | PCDHGA9 | [‘37999834’] | 37999834 |
| 70 | PCDHGA10 | [‘37999843’] | 37999843 |
| 71 | PCDHGA11 | [‘37999842’] | 37999842 |
| 72 | PCDHGA12 | [‘37999478’] | 37999478 |
| 73 | PCDHGB1 | [‘37999833’] | 37999833 |
| 74 | PCDHGB2 | [‘37999832’] | 37999832 |
| 75 | PCDHGB3 | [‘1248281100’] | 1248281100 |
| 76 | PCDHGB4 | [‘37999814’] | 37999814 |
| 77 | PCDHGB5 | [‘37999830’] | 37999830 |
| 78 | PCDHGB6 | [‘37999829’] | 37999829 |
| 79 | PCDHGB7 | [‘37999828’] | 37999828 |
| 83 | PCDHGC3 | [‘37999813’] | 37999813 |
| 84 | PCDHGC4 | [‘37999827’] | 37999827 |
| 85 | PCDHGC5 | [‘37999826’] | 37999826 |
| 86 | DSC1 | [‘223590198’, ‘215274235’] | 223590198 |
| 87 | DSC2 | [‘461968’] | 461968 |
| 88 | DSC3 | [‘116241342’, ‘461968’] | 461968 |
| 89 | DSG1 | [‘292495005’] | 292495005 |
| 90 | DSG2 | [‘148876773’] | 148876773 |
| 91 | DSG3 | [‘239938621’] | 239938621 |
| 92 | DSG4 | [‘60389774’] | 60389774 |
| 93 | CDH13 | [‘1705552’] | 1705552 |
| 94 | CDH26 | [‘1476413386’] | 1476413386 |
| 95 | PCDH1 | [‘215273864’] | 215273864 |
| 96 | PCDH7 | [‘254763320’] | 254763320 |
| 97 | PCDH8 | [‘37999485’] | 37999485 |
| 98 | PCDH9 | [‘206729886’] | 206729886 |
| 99 | PCDH10 | [‘41017507’] | 41017507 |
| 100 | PCDH11X | [‘74761344’] | 74761344 |
| 101 | PCDH11Y | [‘74762719’] | 74762719 |
| 102 | PCDH12 | [‘22095989’] | 22095989 |
| 103 | PCDH17 | [‘118572683’] | 118572683 |
| 104 | PCDH18 | [‘212276496’] | 212276496 |
| 105 | PCDH19 | [‘73620979’] | 73620979 |
| 106 | PCDH20 | [‘544584765’] | 544584765 |
| 107 | CDH1 | [‘399166’, ‘37537753’] | 399166 |
| 108 | CDH2 | [‘116241277’] | 116241277 |
| 109 | CDH3 | [‘146345382’, ‘1705553’] | 146345382 |
| 110 | CDH4 | [‘81175161’] | 81175161 |
| 111 | CDH15 | [‘1705553’] | 1705553 |
| 112 | CDH5 | [‘322510142’] | 322510142 |
| 113 | CDH6 | [‘1705545’] | 1705545 |
| 114 | CDH7 | [‘296434420’] | 296434420 |
| 115 | CDH8 | [‘13432121’] | 13432121 |
| 116 | CDH9 | [‘119370307’] | 119370307 |
| 117 | CDH10 | [‘116241276’] | 116241276 |
| 118 | CDH11 | [‘146345381’] | 146345381 |
| 119 | CDH12 | [‘158937438’] | 158937438 |
| 120 | CDH18 | [‘3023435’] | 3023435 |
| 121 | CDH19 | [‘17366818’, ‘20139065’] | 20139065 |
| 122 | CDH20 | [‘190359622’, ‘37999814’] | 37999814 |
| 123 | CDH22 | [‘24211543’] | 24211543 |
| 124 | CDH24 | [‘38257450’] | 38257450 |
| HNGC ID | Number of variants | Accession numbers |
|---|---|---|
| RET | 20 | [[‘NM_001406778’], [‘NM_001406762’], [‘NM_001406766’], [‘NM_001406780’], [‘NM_001406779’], [‘NM_001406788’], [‘NM_001406774’], [‘NM_001406771’], [‘NM_001406763’], [‘NM_001406768’], [‘NM_001406784’], [‘NM_001406769’], [‘NM_001406787’], [‘NM_001406783’], [‘NM_001406770’], [‘NM_001406760’], [‘NM_001406777’], [‘NM_001406772’], [‘NM_001406776’], [‘NM_001406767’]] |
| PCDHGCT | 0 | [] |
| PCDHGC5 | 2 | [[‘NM_018929’], [‘NM_032407’]] |
| PCDHGC4 | 3 | [[‘NM_001386884’], [‘NM_018928’], [‘NM_032406’]] |
| PCDHGC3 | 3 | [[‘NM_032402’], [‘NM_032403’], [‘NM_002588’]] |
| PCDHGB9P | 0 | [] |
| PCDHGB8P | 0 | [] |
| PCDHGB7 | 2 | [[‘NM_032101’], [‘NM_018927’]] |
| PCDHGB6 | 3 | [[‘NM_018926’], [‘NM_001386906’], [‘NM_032100’]] |
| PCDHGB5 | 2 | [[‘NM_018925’], [‘NM_032099’]] |
| PCDHGB4 | 2 | [[‘NM_003736’], [‘NM_032098’]] |
| PCDHGB3 | 2 | [[‘NM_032097’], [‘NM_018924’]] |
| PCDHGB2 | 2 | [[‘NM_018923’], [‘NM_032096’]] |
| PCDHGB1 | 2 | [[‘NM_018922’], [‘NM_032095’]] |
| PCDHGA9 | 2 | [[‘NM_032089’], [‘NM_018921’]] |
| PCDHGA8 | 2 | [[‘NM_014004’], [‘NM_032088’]] |
| PCDHGA7 | 2 | [[‘NM_032087’], [‘NM_018920’]] |
| PCDHGA6 | 2 | [[‘NM_032086’], [‘NM_018919’]] |
| PCDHGA5 | 2 | [[‘NM_018918’], [‘NM_032054’]] |
| PCDHGA4 | 2 | [[‘NM_032053’], [‘NM_018917’]] |
| PCDHGA3 | 2 | [[‘NM_032011’], [‘NM_018916’]] |
| PCDHGA2 | 2 | [[‘NM_032009’], [‘NM_018915’]] |
| PCDHGA12 | 2 | [[‘NM_032094’], [‘NM_003735’]] |
| PCDHGA11 | 3 | [[‘NM_032091’], [‘NM_032092’], [‘NM_018914’]] |
| PCDHGA10 | 2 | [[‘NM_018913’], [‘NM_032090’]] |
| PCDHGA1 | 2 | [[‘NM_018912’], [‘NM_031993’]] |
| PCDHG@ | 0 | [] |
| PCDHB@ | 0 | [] |
| PCDHB9 | 0 | [] |
| PCDHB8 | 0 | [] |
| PCDHB7 | 0 | [] |
| PCDHB6 | 2 | [[‘NM_018939’], [‘NM_001303145’]] |
| PCDHB5 | 0 | [] |
| PCDHB4 | 0 | [] |
| PCDHB3 | 0 | [] |
| PCDHB2 | 0 | [] |
| PCDHB19P | 0 | [] |
| PCDHB18P | 0 | [] |
| PCDHB17P | 0 | [] |
| PCDHB16 | 0 | [] |
| PCDHB15 | 0 | [] |
| PCDHB14 | 0 | [] |
| PCDHB13 | 0 | [] |
| PCDHB12 | 0 | [] |
| PCDHB11 | 0 | [] |
| PCDHB10 | 0 | [] |
| PCDHB1 | 0 | [] |
| PCDHACT | 0 | [] |
| PCDHAC2 | 2 | [[‘NM_018899’], [‘NM_031883’]] |
| PCDHAC1 | 2 | [[‘NM_018898’], [‘NM_031882’]] |
| PCDHA@ | 0 | [] |
| PCDHA9 | 2 | [[‘NM_031857’], [‘NM_014005’]] |
| PCDHA8 | 2 | [[‘NM_031856’], [‘NM_018911’]] |
| PCDHA7 | 2 | [[‘NM_031852’], [‘NM_018910’]] |
| PCDHA6 | 3 | [[‘NM_031848’], [‘NM_031849’], [‘NM_018909’, ‘XM_001124723’, ‘XM_001124826’]] |
| PCDHA5 | 2 | [[‘NM_031501’], [‘NM_018908’]] |
| PCDHA4 | 2 | [[‘NM_031500’], [‘NM_018907’]] |
| PCDHA3 | 2 | [[‘NM_031497’], [‘NM_018906’]] |
| PCDHA2 | 3 | [[‘NM_031495’], [‘NM_031496’, ‘XM_005268491’, ‘XM_005278372’], [‘NM_018905’]] |
| PCDHA14 | 0 | [] |
| PCDHA13 | 2 | [[‘NM_018904’], [‘NM_031865’]] |
| PCDHA12 | 2 | [[‘NM_031864’], [‘NM_018903’]] |
| PCDHA11 | 2 | [[‘NM_018902’], [‘NM_031861’]] |
| PCDHA10 | 3 | [[‘NM_031859’], [‘NM_031860’], [‘NM_018901’]] |
| PCDHA1 | 3 | [[‘NM_031411’], [‘NM_031410’], [‘NM_018900’]] |
| PCDH9 | 15 | [[‘XM_054374595’], [‘XM_054374594’], [‘XM_054374593’], [‘XM_054374592’], [‘XM_054374591’], [‘XM_017020621’], [‘XM_005266408’], [‘XM_017020620’], [‘XM_017020619’], [‘XM_011535099’], [‘NM_001318374’], [‘NM_020403’], [‘NM_001318372’, ‘XM_005266406’], [‘NM_001318373’, ‘XM_005266407’], [‘NM_203487’]] |
| PCDH8 | 2 | [[‘NM_032949’], [‘NM_002590’]] |
| PCDH7 | 3 | [[‘NM_002589’], [‘NM_001173523’], [‘NM_032457’]] |
| PCDH20 | 1 | [[‘XM_054374865’]] |
| PCDH19 | 3 | [[‘NM_001105243’], [‘NM_020766’, ‘XM_033173’], [‘NM_001184880’]] |
| PCDH18 | 6 | [[‘XM_054350249’], [‘XM_054350248’], [‘XM_017008311’], [‘XM_006714239’], [‘NM_001300828’, ‘XM_005263070’], [‘NM_019035’]] |
| PCDH17 | 6 | [[‘XM_054374468’], [‘XM_054374467’], [‘XM_054374466’], [‘XM_017020547’], [‘XM_047430276’], [‘XM_005266357’]] |
| PCDH15 | 20 | [[‘NM_001142767’], [‘NM_001142770’], [‘NM_001142773’], [‘NM_001142764’], [‘NM_001142772’], [‘NM_033056’, ‘XM_373461’, ‘XM_937929’], [‘NM_001354420’, ‘XM_017016572’], [‘NM_001354429’, ‘XM_017016571’], [‘NM_001142765’], [‘NM_001142763’], [‘NM_001354430’], [‘NM_001142766’], [‘NM_001354411’], [‘NM_001142768’], [‘NM_001142769’], [‘NM_001142771’], [‘NM_001384140’], [‘XM_054366621’], [‘XM_054366620’], [‘XM_047425664’]] |
| PCDH12 | 1 | [[‘XM_054352749’]] |
| PCDH11Y | 11 | [[‘XM_054328381’], [‘XM_054328380’], [‘XM_054328379’], [‘XM_017030081’], [‘XM_017030080’], [‘XM_017030079’], [‘NM_001395587’], [‘NM_032972’], [‘NM_001278619’], [‘NM_032971’], [‘NM_032973’]] |
| PCDH11X | 20 | [[‘XM_054326845’], [‘XM_054326844’], [‘XM_054326843’], [‘XM_054326842’], [‘XM_054326841’], [‘XM_054326840’], [‘XM_054326839’], [‘XM_054326838’], [‘XM_047441995’], [‘XM_017029421’], [‘XM_017029419’], [‘XM_011530914’], [‘XM_047441994’], [‘XM_047441993’], [‘XM_011530911’], [‘XM_017029416’], [‘XM_011530910’], [‘NM_032968’], [‘NM_032969’], [‘NM_001168363’]] |
| PCDH10 | 4 | [[‘NM_020815’], [‘NM_032961’], [‘XM_054350597’], [‘XM_011532150’]] |
| PCDH1 | 11 | [[‘XM_054352708’], [‘XM_054352707’], [‘XM_054352706’], [‘XM_054352705’], [‘XM_017009517’], [‘XM_005268454’], [‘XM_005268452’], [‘NM_032420’], [‘NM_002587’], [‘NM_001278613’], [‘NM_001278615’]] |
| FAT4 | 7 | [[‘XM_054350821’], [‘XM_011532237’], [‘XM_047416153’], [‘XM_047416151’], [‘NM_001291285’, ‘XM_005263210’], [‘NM_024582’], [‘NM_001291303’, ‘XM_006714304’, ‘XM_011532236’]] |
| FAT3 | 16 | [[‘XM_054367644’], [‘XM_054367643’], [‘XM_054367642’], [‘XM_054367641’], [‘XM_017017187’], [‘XM_017017186’], [‘XM_017017185’], [‘XM_017017184’], [‘XM_017017182’], [‘XM_017017180’], [‘XM_047426353’], [‘XM_017017179’], [‘XM_017017178’], [‘NM_001378141’], [‘NM_001008781’, ‘XM_017017183’, ‘XM_061871’], [‘NM_001367949’, ‘XM_017017181’]] |
| FAT2 | 12 | [[‘XM_054352053’], [‘XM_054352052’], [‘XM_054352051’], [‘XM_054352050’], [‘XM_054352049’], [‘XM_054352048’], [‘XM_017009227’], [‘XM_047416934’], [‘XM_017009225’], [‘XM_017009224’], [‘XM_011537600’], [‘XM_011537603’]] |
| FAT1 | 6 | [[‘XM_054349259’], [‘XM_054349258’], [‘XM_054349257’], [‘XM_006714139’], [‘XM_005262835’], [‘XM_005262834’]] |
| DSG4 | 2 | [[‘NM_001134453’], [‘NM_177986’]] |
| DSG3 | 2 | [[‘XM_054318238’], [‘XM_011525850’]] |
| DSG2 | 2 | [[‘XM_054318237’], [‘XM_047437315’]] |
| DSG1 | 0 | [] |
| DSC3 | 2 | [[‘NM_024423’], [‘NM_001941’]] |
| DSC2 | 4 | [[‘NM_001406506’, ‘XM_005258206’], [‘NM_001406507’], [‘NM_004949’], [‘NM_024422’]] |
| DSC1 | 3 | [[‘XM_054318236’], [‘NM_024421’], [‘NM_004948’]] |
| DCHS2 | 3 | [[‘NM_001412223’], [‘NM_001142552’], [‘NM_001358235’]] |
| DCHS1 | 0 | [] |
| CLSTN3 | 7 | [[‘XM_054373906’], [‘XM_054373905’], [‘XM_054332547’], [‘XM_054332546’], [‘XM_047429921’], [‘XM_047429920’], [‘XM_047429919’]] |
| CLSTN2 | 2 | [[‘XM_054347514’], [‘XM_017007022’]] |
| CLSTN1 | 5 | [[‘NM_001009566’, ‘XM_937951’], [‘NM_014944’], [‘NM_001302883’, ‘XM_005263432’], [‘XM_054335191’], [‘XM_047449470’]] |
| CELSR3 | 0 | [] |
| CELSR2 | 0 | [] |
| CELSR1 | 10 | [[‘NM_014246’], [‘NM_001378328’, ‘XM_006724383’], [‘XM_054326177’], [‘XM_054326176’], [‘XM_054326175’], [‘XM_054326174’], [‘XM_011530555’], [‘XM_011530554’], [‘XM_011530553’], [‘XM_047441624’]] |
| CDHR5 | 18 | [[‘XM_054369143’], [‘XM_054369142’], [‘XM_054369141’], [‘XM_054369140’], [‘XM_054369139’], [‘XM_054328908’], [‘XM_054328907’], [‘XM_054328906’], [‘XM_054328905’], [‘XM_054328904’], [‘XM_011520190’], [‘XM_024448584’], [‘XM_011520189’], [‘XM_011520188’], [‘XM_006718253’], [‘NM_001171968’], [‘NM_031264’], [‘NM_021924’]] |
| CDHR4 | 16 | [[‘XM_054346474’], [‘XM_054346473’], [‘XM_054346472’], [‘XM_054346471’], [‘XM_054346470’], [‘XM_054346469’], [‘XM_054346468’], [‘XM_054346467’], [‘XM_011533701’], [‘XM_011533700’], [‘XM_017006371’], [‘XM_017006370’], [‘XM_017006369’], [‘XM_017006368’], [‘XM_017006367’], [‘XM_017006366’]] |
| CDHR3 | 2 | [[‘NM_001301161’, ‘XM_005250223’], [‘NM_152750’, ‘XR_108861’, ‘XR_110727’, ‘XR_114075’]] |
| CDHR2 | 2 | [[‘NM_017675’], [‘NM_001171976’]] |
| CDHR1 | 12 | [[‘XM_054367149’], [‘XM_054367148’], [‘XM_054367147’], [‘XM_054367146’], [‘XM_054367145’], [‘XM_047425997’], [‘XM_011540340’], [‘XM_011540339’], [‘XM_011540338’], [‘XM_011540337’], [‘NM_001171971’], [‘NM_033100’]] |
| CDH9 | 0 | [] |
| CDH8 | 8 | [[‘XM_054379349’], [‘XM_054379348’], [‘XM_054379347’], [‘XM_047433484’], [‘XM_047433482’], [‘XM_005255760’], [‘NM_001410893’, ‘XM_047433483’], [‘NM_001796’]] |
| CDH7 | 4 | [[‘NM_033646’], [‘NM_001362438’, ‘XM_017025523’], [‘NM_001317214’], [‘NM_004361’]] |
| CDH6 | 7 | [[‘XM_054351377’], [‘XM_054351376’], [‘XM_054351375’], [‘XM_047416591’], [‘XM_011513921’], [‘NM_001362435’, ‘XM_017008911’], [‘NM_004932’]] |
| CDH5 | 8 | [[‘XM_054379336’], [‘XM_054379335’], [‘XM_054379334’], [‘XM_054379333’], [‘XM_047433471’], [‘XM_047433470’], [‘XM_047433469’], [‘XM_011522801’]] |
| CDH4 | 7 | [[‘NM_001252339’], [‘NM_001794’, ‘NM_024883’, ‘XM_372872’], [‘NM_001252338’], [‘XM_054322813’], [‘XM_054322812’], [‘XM_047439813’], [‘XM_047439812’]] |
| CDH3 | 7 | [[‘XM_054379318’], [‘XM_054379317’], [‘XM_047433450’], [‘XM_011522800’], [‘NM_001317195’], [‘NM_001317196’], [‘NM_001793’]] |
| CDH26 | 5 | [[‘NM_001348204’], [‘NM_021810’], [‘NM_177980’], [‘XM_054323793’], [‘XM_011528970’]] |
| CDH24 | 6 | [[‘XM_054376605’], [‘XM_054376604’], [‘XM_047431697’], [‘XM_011537089’], [‘NM_144985’], [‘NM_022478’]] |
| CDH23 | 9 | [[‘NM_001171932’], [‘NM_001171930’], [‘NM_052836’], [‘NM_001171931’], [‘NM_022124’], [‘NM_001171936’], [‘NM_001171935’], [‘NM_001171934’], [‘NM_001171933’]] |
| CDH22 | 10 | [[‘XM_054323846’], [‘XM_054323845’], [‘XM_054323844’], [‘XM_054323843’], [‘XM_054323842’], [‘XM_047440374’], [‘XM_024451967’], [‘XM_024451966’], [‘XM_047440373’], [‘XM_011528994’]] |
| CDH20 | 2 | [[‘XM_054318533’], [‘XM_024451165’]] |
| CDH2 | 5 | [[‘NM_001792’], [‘NM_001308176’, ‘XM_005258182’], [‘XM_054318138’], [‘XM_017025514’], [‘XM_011525788’]] |
| CDH19 | 10 | [[‘XM_054318543’], [‘XM_054318542’], [‘XM_054318541’], [‘XM_054318540’], [‘XM_047437485’], [‘XM_011525932’], [‘XM_011525931’], [‘XM_047437484’], [‘NM_001271028’], [‘NM_021153’, ‘XM_005266688’]] |
| CDH18 | 20 | [[‘NM_001349563’, ‘XM_017008933’], [‘NM_001349558’, ‘XM_017008932’], [‘NM_001167667’], [‘NM_001349562’, ‘XM_017008934’], [‘NM_001349556’, ‘XM_017008925’], [‘NM_001349559’, ‘XM_017008931’], [‘NM_001291956’], [‘NM_001349561’, ‘XM_017008941’], [‘NM_001349560’, ‘XM_017008937’], [‘NM_004934’], [‘NM_001291957’, ‘XM_006714437’], [‘XM_054351396’], [‘XM_054351395’], [‘XM_054351394’], [‘XM_054351393’], [‘XM_054351392’], [‘XM_054351391’], [‘XM_054351390’], [‘XM_054351389’], [‘XM_054351388’]] |
| CDH17 | 13 | [[‘NM_001413961’], [‘NM_001413959’, ‘XM_011516790’], [‘NM_001413958’], [‘NM_001413957’], [‘NM_001413956’], [‘NM_001413955’], [‘NM_001413954’], [‘NM_001413953’], [‘NM_001144663’], [‘NM_004063’], [‘NM_001413960’], [‘NM_001413952’], [‘NM_001413951’]] |
| CDH16 | 10 | [[‘XM_054379375’], [‘XM_054379374’], [‘XM_054379373’], [‘XM_005255770’], [‘XM_047433490’], [‘XM_011522807’], [‘NM_001204746’], [‘NM_001204744’], [‘NM_001204745’], [‘NM_004062’]] |
| CDH15 | 0 | [] |
| CDH13 | 12 | [[‘XM_054379358’], [‘XM_054379357’], [‘XM_054379356’], [‘XM_017022849’], [‘XM_017022848’], [‘XM_011522804’], [‘NM_001220492’], [‘NM_001220490’], [‘NM_001220488’], [‘NM_001220491’], [‘NM_001220489’], [‘NM_001257’]] |
| CDH12 | 11 | [[‘XM_054351385’], [‘XM_047416602’], [‘NM_001317227’], [‘NM_001364108’], [‘NM_001364107’, ‘XM_011513927’], [‘NM_001364109’], [‘NM_001364106’, ‘XM_017008921’], [‘NM_001364104’, ‘XM_017008920’], [‘NM_001364105’], [‘NM_004061’], [‘NM_001317228’]] |
| CDH11 | 7 | [[‘XM_054379352’], [‘XM_054379351’], [‘XM_054379350’], [‘XM_047433486’], [‘NM_001330576’, ‘XM_017022830’], [‘NM_001308392’, ‘XM_005255764’], [‘NM_001797’]] |
| CDH10 | 6 | [[‘XM_054351383’], [‘XM_011513923’], [‘NM_001317222’], [‘NM_006727’], [‘NM_001317224’, ‘XM_011513925’, ‘XM_017008915’], [‘NM_001362460’]] |
| CDH1 | 4 | [[‘NM_001317185’], [‘NM_001317186’], [‘NM_001317184’], [‘NM_004360’]] |